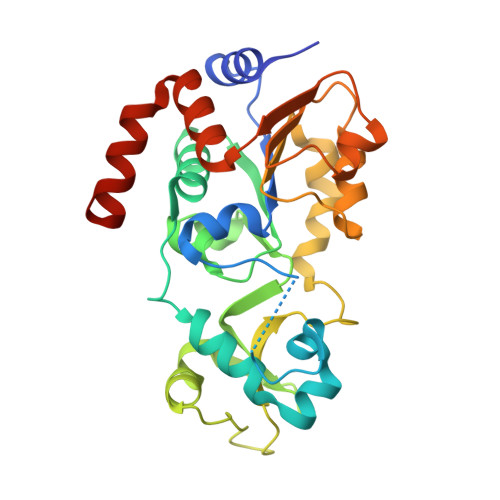
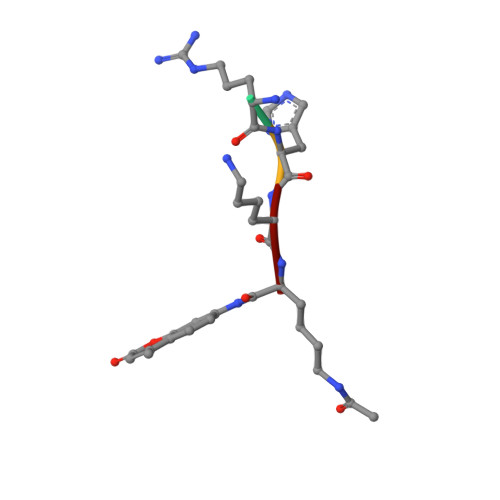
Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Nguyen, G.T.T., Gertz, M., Steegborn, C.(2013) Chem Biol 20: 1375
- PubMed: 24211137
- DOI: https://doi.org/10.1016/j.chembiol.2013.09.019
- Primary Citation of Related Structures:
4C78, 4C7B - PubMed Abstract:
Sirtuins are protein deacetylases regulating aging processes and various physiological functions. Resveratrol, a polyphenol found in red wine, activates human Sirt1 and inhibits Sirt3, and it can mimic calorie restriction effects, such as lifespan extension in lower organisms. The mechanism of Sirtuin modulation by resveratrol is not well understood. We used 4'-bromo-resveratrol (5-(2-(4-hydroxyphenyl)vinyl)-1,3-benzenediol) to study Sirt1 and Sirt3 modulation. Despite its similarity to the Sirt1 activator resveratrol, the compound potently inhibited both, Sirt1 and Sirt3. Crystal structures of Sirt3 in complex with a fluorophore-labeled and with a native substrate peptide, respectively, in presence of 4'-bromo-resveratrol reveal two compound binding sites. Biochemical studies identify the internal site and substrate competition as the mechanism for inhibition, providing a drug target site, and homology modeling suggests that the second, allosteric site might indicate the site for Sirt1 activation.
Organizational Affiliation:
Department of Biochemistry, University of Bayreuth, 95440 Bayreuth, Germany.